Vibe Check Your Geospatial Foundation Model
with geovibes!
TLDR;
Using Geovibes you can interact with a foundation model/embeddings using a jupyter notebook via similarity searching on your laptop.
This gives you a sense of your model’s vibes, i.e minutiae in the data/minority classes that perhaps only you care about, and aren’t measured in benchmarks.
We provide a script that enables you to build a searchable index from embeddings provided online.
We suggest the community uses the following schema for the release of embeddings: geoparquet files with [tile_id, geometry, embedding] where geometry is a (lat, lon) representing the centre of a tile. Each geoparquet file should contain the embeddings generated over an Military Grid Reference System (MGRS) tile.
Metadata on temporal range, tile size, overlap and pixel resolution should also be included
Potential future plans may involve: dealing with embedding rasters, making this more memory efficient, an embeddings pipeline for you to generate your own using custom models, experiments showing that this is useful.
Why?
I feel like earth observation research is currently a very `vibes-based` community.
Expensive foundation models get released without benchmarks. Global embedding datasets get released with limited benchmarking on downstream tasks. The benchmarks that do exist are narrow in geographic scope, and some are already maxed out by models that we know are just not that great. There also appears to be a general lack of procedure around benchmarking: some papers might benchmark on PANGAEA, others on Geo-Bench, and others on their own collection of tasks.
Most of us don’t read the actual papers released, we just go off the LinkedIn posts and the vibes. Myself included!
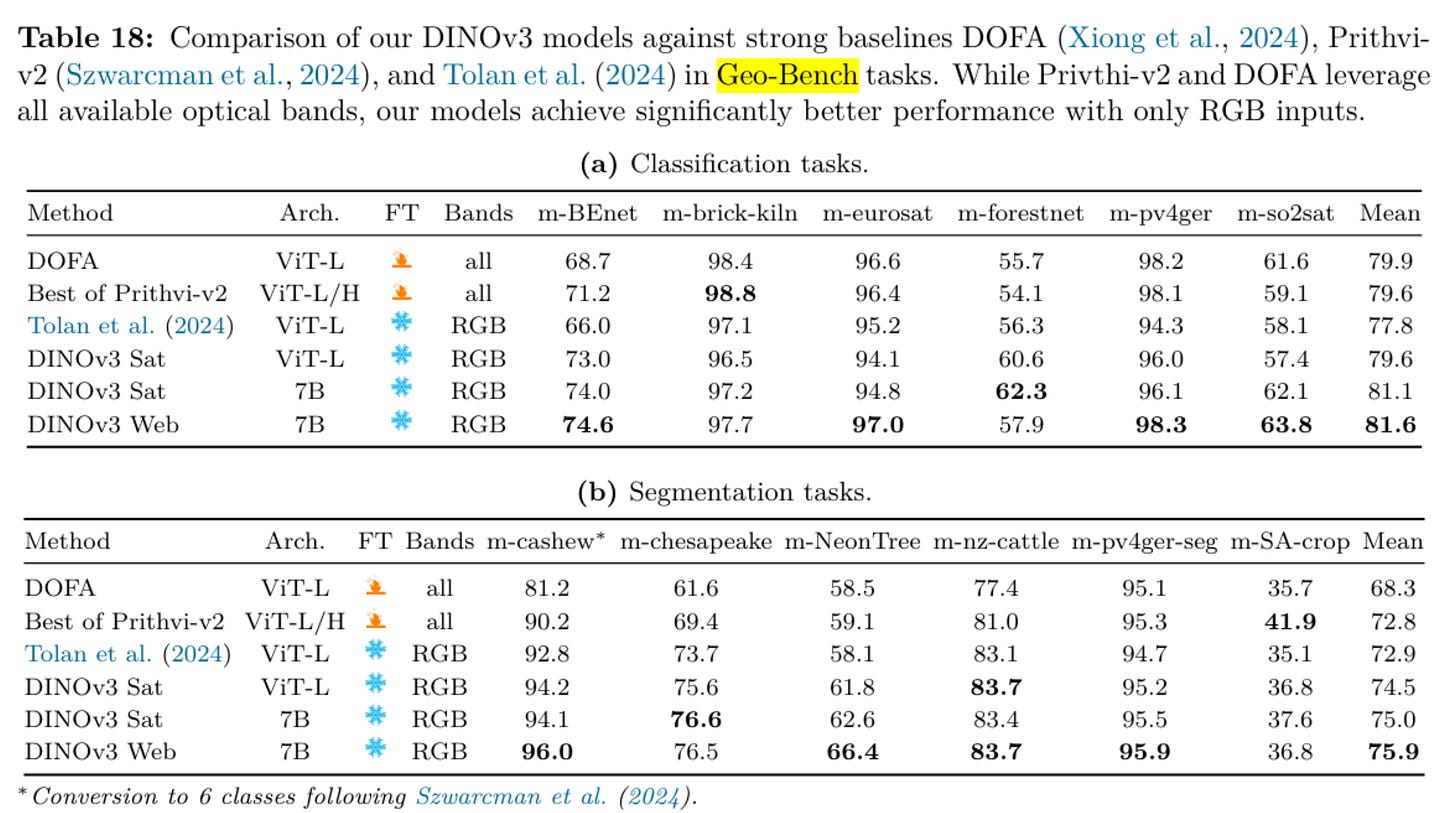
As an example just look at this set of tables from the DINOv3 paper: m-brick-kiln, m-eurosat, m-pv4ger, m-pv4ger-seg or 4/12 of these tasks are clearly saturated across all of the models being compared here. These should be taken out to pasture.
My feeling is that these ‘general’ sets of benchmarks targeted at measuring how foundational a given model is don’t reflect what users are actually interested in. We can acknowledge that those in academia must play the numbers game and over-index on novelty and benchmark performance as opposed to impact and utility (I have been guilty of this myself), but that does not mean that the rest of us should follow suit!
We see the same effect with LLMs coding agents: frontier labs will release benchmark results, but the thing I tend to pay attention to are real users vibe checks. After all, these are real users, not static benchmarks that can be gamed! The goal of geovibes is to provide a framework for users to vibe check embeddings/foundation models easily.
How Do I Use It?
Geovibes is a weekend project. It should not be used as a production tool. I’m not even sure how you would do that to be honest. There are probably a million small things that are broken, and some big ones. With that being said: you should use geovibes to create datasets and maps.
A pretty simple, universal workflow for mapping is:
label positives and negatives with Geovibes
you may need to sample more diverse negative samples since your dataset will only consist of ‘hard’ ones resulting from the search. Landcover is a great way to do this.
train a classifier (XGBoost/Random Forest/LightGBM/Catboost) on top of those embeddings.
Once you have a trained classifier: deploy this classifier over any set of embeddings generated by the same source model.
The classification workflow has not been implemented as of yet in geovibes, but you can subscribe to updates to be notified when it is.
What is Geovibes?
We built on top of the open source ei-notebook released by the Earth Genome to address this issue: if we’re going to be vibes-based, then let’s do it for real! Yes, we are missing a strong set of universal benchmarks in EO, but then again is that really important? Each user has their own specific use-case they care about: we should instead empower them to easily get a ‘feel’ for a given set of embeddings aka the vibes.
Let’s allow the user to interact iteratively with these embeddings, the same way they would do with chatbots/coding agents to figure out if they prefer Gemini Pro 2.5, Opus 4.1, GPT-5-Codex or GPT-5-High or … you get the idea.
Geovibes is a tool that allows you to create or download local, searchable vector databases of embeddings produced by earth observation foundation models. It comes at a time when we have an increasing availability of embedding datasets (Earth Genome Softcon, Google Alpha Earth, TESSERA etc…), but no unified or easy way to interact with them.
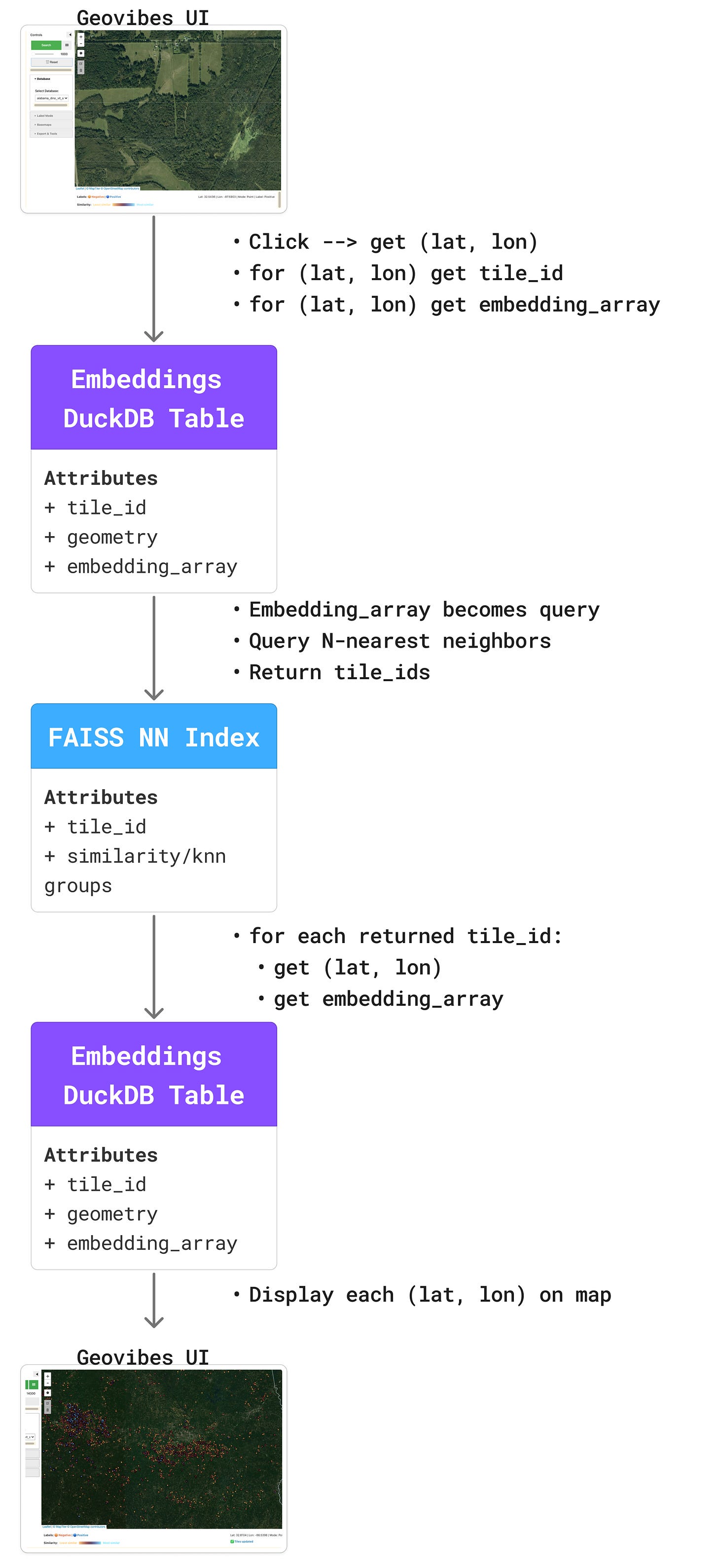
Think of it as Earth Index, but it runs on your laptop. This allows you to checkout the vibes of any new embedding dataset or foundation model. The idea is relatively simple: click on a location, retrieve the embedding vector for that location, use it to query your database using similarity search, then return the top-k neighbors.
Human in the Loop Searching
You needn’t stop there though: what if the returned search results contain some relevant results, but some irrelevant ones? Geovibes enables you to label negative samples: essentially pushing your query vector to be more different than some selected points.
In the video below: I’m using Google’s Alpha Earth Foundations (AEF) embeddings in an attempt to find aquaculture in Alabama. I click on an example and zoom out: I see that because AEF embeddings do not contain any spatial information, it seems that both the Tombigbee River (West) and the Alabama River (East) are picked out by my search results (compare the image below and the video).
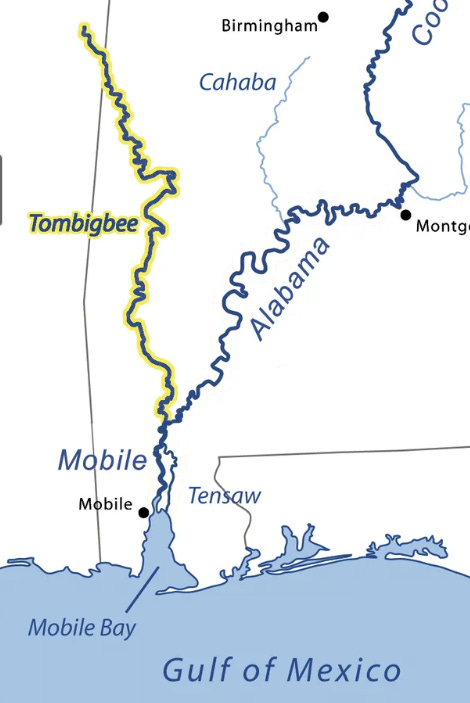
These are clearly not the type of thing I am looking for, and I can indicate this to my search: I draw a polygon around the Tombigbee River search results, add these as negatives to my dataset, and perform a new search with my updated query vector. We see that many search results from the Alabama River disappear as a result of this, even though I did not label those as negatives!
This type of operation is valuable for two reasons: it makes the similarity searching steerable to some degree AND it helps surface ‘hard negatives’ which are useful for training classifiers from your dataset.
How it Works
I want to specifically credit Ed Boyda from the Earth Genome with coming up with the idea to run this type of search locally: as far as I’m aware (correct me if I’m wrong Earth Genies), he implemented the original version of this which used annoy and held the embeddings in a GeoDataFrame in memory.
The ‘backend’ of geovibes relies on two core technologies: faiss for similarity searching, and duckdb for fast + local access to embeddings. Using the duckdb vss extension, you can technically do this without faiss, but we found that solution scaled poorly both in terms of index-build time, and search time.
Given a set of embedding vectors: we build a faiss index over the embeddings, this tells us for a given vector, which vectors are similar for all vectors in the dataset.
We link this faiss index to a duckdb table by
tile_id: the faiss index returns which tile IDs are most similar to a given query,We then use duckdb to retrieve the embedding vectors for those tile IDs. This enables us to re-calculate and update a given query vector with the actual embedding values, as well as display them on the UI for the user to peruse.
For now: you can use geovibes to checkout embeddings and foundation models. Which ones are useful for your specific use cases? Are you able to find your things immediately, or does it take several iterations of labeling?
Available Searchable Databases
We have pre-compiled some faiss indexes + duckdb tables for you to check out and play around with. For now, searchable indexes are available over Alabama and New Mexico. These are hosted on Source Cooperative (thanks Jed!).
Alabama:
Earth Genome SSL4EO DINO ViT Sentinel-2 embeddings 2024-2025
Earth Genome Quantized SSL4EO DINO ViT Sentinel-2 embeddings 2024-2025
Earth Genome Quantized Softcon embeddings 2024-2025
Google Satellite Embeddings v1 2024-2025
Clay v1.5 NAIP embeddings 2019-2020 (via a heroic effort from Zoe, thank you!)
New Mexico:
Google Satellite Embeddings v1 2024-2025
Please get in touch if you’d like us to generate other regions.
You can access these via the download_embeddings.py script in geovibes (thanks Noah!): if you don’t have a great internet connection, I recommend you start with a Google Satellite Embedding v1 database, as these are only 64-dimensional and were created with no tile-overlap, so are much smaller. The Clay v1.5 database is about 30 GB (!!).
Bring Your Own Embeddings
We provide scripts in geovibes such that users can generate their own searchable indexes which are compatible with the geovibes user interface. In order for these to work, your embeddings should follow this schema: [tile_id, geometry, embedding] where tile_id is a unique identifier, geometry is a (lat, lon) representing the centre of a tile and embedding is the embedding array.
We suggest that if others would like to make open tiled embedding datasets, that use the geoparquet format, separate out embeddings by MGRS tile, and use this schema. Additionally, metadata such as temporal range, tile size, overlap and pixel resolution should also be included in the filenames/folder structure.
Our hope is that the this tool increases utilization of some already existing embedding datasets, and encourages others to release more datasets knowing that they can now be searched over fairly large regions on a laptop.
What’s Next?
Many embeddings appear to be released as rasters currently, at the pixel level. I think it’d be interesting to see how/if we can support those in this kind of workflow.
We’ll be writing some posts about fun problems we encountered while doing this:
including how duckdb’s vss extension doesn’t appear to scale well,
why we think smaller embeddings are better,
how duckdb/faiss appear to scale with search size/dataset size/request size,
how far we think we can push this kind of local workflow on our laptops.
We’ll also be using the tool to do some mapping, and probably finding lots of bugs along the way, you can expect us to update the tool as we use it, but you should feel free to fork it and modify it to fit your own purposes as well.
With LLMs/coding agents as they are: there is no need to have a ‘central’ repository. If you want to modify it to fit your purposes, or fix a bug, just ask Codex or Claude to do it, no need to submit a PR/github issue unless you really want to.
There is more than one way to skin a foundation model.
Acknowledgements
Thank you to the Earth Genome, in particular Ben Strong, Hutch Ingold, and Ed Boyda for supporting this effort by providing embeddings and agreeing to open source the original ei-notebook implementation.
Thank you to Zoe, Noah and Isaac who contributed to Geovibes, and listened to me rant about foundation models and embeddings.
Thank you to Jed Sundwall, who enabled us to distribute these databases via Source Cooperative.